A new paper at PLoS ONE featuring ancient mitochondrial (mtDNA) data from Wielbark, Przeworsk and early Slavic remains argues for matrilineal continuity in present-day Poland since the Iron Age. It's actually based on a thesis that I blogged about more than two years ago (see here). However, it does include some fresh insights, so it's worth a look even if you read the thesis. RoIA stands for Roman Iron Age.
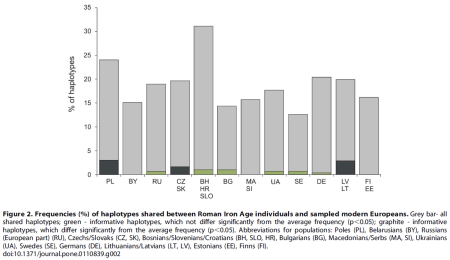
Three modern populations or groups of populations (Lithuanians and Latvians, Poles, and Czechs and Slovaks) were found to contain significantly higher percentages (p,0.05) of shared informative haplotypes with the RoIA samples compared to other present-day populations (Figure 2, Table S4). Notably, modern Poles shared the highest number (nine) of informative mtDNA haplotypes with the RoIA individuals.
...
Of particular interest are three RoIA samples assigned to subhaplogroup H5a1, which were recovered from the Kowalewko (sample K1), the Gaski, and the Rogowo (samples G1 and R3) burial sites (see Figure 1). Recent studies on mtDNA hg H5 have revealed that phylogenetically older subbranches, H5a3, H5a4 and H5e, are observed primarily in modern populations from southern Europe, while the younger ones, including H5a1 that was found among RoIA individuals in our study, date to around 4.000 years ago (kya) and are found predominantly among Slavic populations of Central and East Europe, including contemporary Poles [15]. Notably, we also found one ME sample belonging to subhaplogroup H5a1 (sample OL1 in Table 3). The presence of subclusters of H5a1 in four ancient samples belonging to both the RoIA and the ME periods, and in contemporary Poles, indicates the genetic continuity of this maternal lineage in the territory of modern-day Poland from at least Roman Iron Age i.e., 2 kya.
...
The evolutionary age of H5 sub-branches (,4 kya) [15] also approximates the age of N1a1a2 subclade found in the RoIA population (sample KA2) (Table 2). The coalescence age of N1a1a2 is around 3.4–4 kya, making this haplotype one of the youngest sub-branches within hg N [52]. The N1a1a2 haplotype found in one RoIA individual was classified as unique because no exact match was found among the twelve comparative populations or groups of populations used in the haplotype sharing test. Notably, a similar N1a1a2 haplotype carrying an additional transition at position 16172 was found in a modern-day Polish individual [53].
I suspect the publication of these results at this time, so many months after they were first revealed in the aforementioned thesis, is part of an effort to drum up interest and secure funding for a new project on the genetic history of Greater Poland, which was announced late last year (see here). I say that because one of the people organizing the project, Janusz Piontek, is also listed as a co-author on this paper. So if we're lucky we might soon see full genome sequences from a few of these Iron Age and Medieval samples.
Citation...
Juras A, Dabert M, Kushniarevich A, Malmstro¨m H, Raghavan M, et al. (2014) Ancient DNA Reveals Matrilineal Continuity in Present-Day Poland over the Last Two Millennia. PLoS ONE 9(10): e110839. doi:10.1371/journal.pone.0110839
A new paper in the Economics & Human Biology journal argues that male height in Europe is mostly determined by nutrition and genetics. That's not exactly earth shattering news. However, the authors also point out that Y-chromosome haplogroup I-M170 shows a strong correlation with the highest average stature on the continent, and speculate that the link between the two might be Upper Paleolithic hunter-gatherer ancestry:
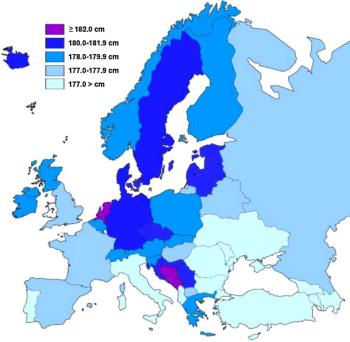
The average height of 45 national samples used in our study was 178.3 cm (median 178.5 cm). The average of 42 European countries was 178.3 cm (median 178.4 cm). When weighted by population size, the average height of a young European male can be estimated at 177.6 cm. The geographical comparison of European samples (Fig. 1) shows that above average stature (178+ cm) is typical for Northern/Central Europe and the Western Balkans (the area of the Dinaric Alps). This agrees with observations of 20th century anthropologists (Coon, 1939; Lundman 1977). At present, the tallest nation in Europe (and also in the world) are the Dutch (average male height 183.8 cm), followed by Montenegrins (183.2 cm) and possibly Bosnians (182.5 cm) (Table 1). In contrast with these high values, the shortest men in Europe can be found in Turkey (173.6 cm), Portugal (173.9 cm), Cyprus (174.6 cm) and in economically underdeveloped nations of the Balkans and former Soviet Union (mainly Albania, Moldova, and the Caucasian republics).
...
The trend of increasing height has already stopped in Norway, Denmark, the Netherlands, Slovakia and Germany. In Norway, military statistics date its cessation to late 1980s.
...
In contrast, the fastest pace of the height increase (≥1 cm/decade) can be observed in Ireland, Portugal, Spain, Latvia, Belarus, Poland, Bosnia and Herzegovina, Croatia, Greece, Turkey and at least in the southern parts of Italy.
...
Although the documented differences in male stature in European nations can largely be explained by nutrition and other exogenous factors, it is remarkable that the picture in Fig. 1 strikingly resembles the distribution of Y haplogroup I-M170 (Fig. 10a). Apart from a regional anomaly in Sardinia (sub-branch I2a1a-M26), this male genetic lineage has two frequency peaks, from which one is located in Scandinavia and northern Germany (I1-M253 and I2a2-M436), and the second one in the Dinaric Alps in Bosnia and Herzegovina (I2a1b-M423)16. In other words, these are exactly the regions that are characterized by unusual tallness. The correlation between the frequency of I-M170 and male height in 43 European countries (including USA) is indeed highly statistically significant (r = 0.65; p < 0.001) (Fig. 11a, Table 4). Furthermore, frequencies of Paleolithic Y haplogroups in Northeastern Europe are improbably low, being distorted by the genetic drift of N1c-M46, a paternal marker of Ugrofinian hunter-gatherers. After the exclusion of N1c-M46 from the genetic profile of the Baltic states and Finland, the r-value would further slightly rise to 0.67 (p < 0.001). These relationships strongly suggest that extraordinary predispositions for tallness were already present in the Upper Paleolithic groups that had once brought this lineage from the Near East to Europe.
Citation...
Grasgruber et al., The role of nutrition and genetics as key determinants of the positive height trend, Economics & Human Biology, available online 7 August 2014, DOI: 10.1016/j.ehb.2014.07.002
There's been a lot of horseshit published over the years about Y-chromosome haplogroup R1a, which just happens to be my haplogroup. This includes academic papers in journals like PLoS ONE and Nature.
Indeed, a new paper on the phylogeography of R1a appeared at the Nature website today: Underhill et al. 2014. It's actually a much better effort than anything else on the topic at academic level thus far, but certainly not without issues.
For instance, the authors failed to include two well known and very important R1a subclades in their analysis: the Northwest European-specific R1a-CTS4385 and the East and Central European-specific R1a-Z280. As a result, the former is lumped with R1a-M417* and the latter with R1a-Z282*. In fact, Z280 is shown to be above Z282 in the topology of R1a-M420 (see Figure 1 here), which is plain wrong. These are major oversights and mean that this study is not a very useful resource as far as the phylogeography of European R1a is concerned.
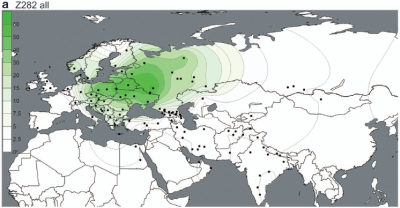
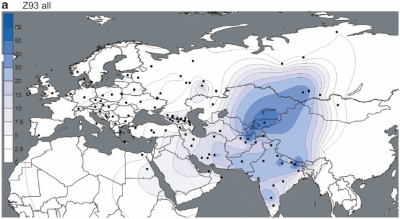
But the paper does show a couple of interesting things. For instance, the maps below offer the best illustration to date of the dichotomy between the European-specific R1a-Z282 and Asian-specific R1a-Z93.
However, these are very closely related subclades, sharing the Z645 mutation (unfortunately not mentioned in the paper), and both reaching high frequencies among Indo-European speakers. It's therefore plausible that groups carrying these markers expanded to the west and east from a zone between their current hotspots, possibly the Volga-Ural region, rather recently.
These migrations had to have happened after 4800-6800 YBP, which is the age of R1a-M417 reported by Underhill et al., and backed up by estimates from genetic genealogists using, among other things, complete R1a sequences (see here). In other words, the rapid expansions of R1a-Z282 and R1a-Z93 appear to have taken place from more or less the same region during the generally accepted early Indo-European timeframe, making them excellent candidates for paternal markers of the early Indo-European dispersals.
At the same time, the paucity of R1a-Z93 and derived lineages in Europe, including Eastern Europe, suggests that historic migrations originating in East and Central Asia, like those of the early Turks, had a negligible effect on the paternal ancestry of modern Europeans. This shows very clearly on the PCA in Figure 4 (see here).
Citation...
Underhill et al., The phylogenetic and geographic structure of Y-chromosome haplogroup R1a, European Journal of Human Genetics, advance online publication, 26 March 2014; doi:10.1038/ejhg.2014.50
See also...
The beast among Y-haplogroups